import striplog
striplog.__version__
'unknown'
Block logs#
We’d like to make blocky, upscaled versions of logs.
Let’s load a well from an LAS File using welly:
from welly import Well
w = Well.from_las('P-129_out.LAS')
w
| Kennetcook #2 Long = 63* 45'24.460 W | |
|---|---|
| crs | CRS({}) |
| location | Lat = 45* 12' 34.237" N |
| country | CA |
| province | Nova Scotia |
| latitude | |
| longitude | |
| datum | |
| section | 45.20 Deg N |
| range | PD 176 |
| township | 63.75 Deg W |
| td | None |
| data | CALI, DPHI_DOL, DPHI_LIM, DPHI_SAN, DRHO, DT, DTS, GR, HCAL, NPHI_DOL, NPHI_LIM, NPHI_SAN, PEF, RHOB, RLA1, RLA2, RLA3, RLA4, RLA5, RM_HRLT, RT_HRLT, RXOZ, RXO_HRLT, SP |
gr = w.data['GR']
gr
| GR [gAPI] | |
|---|---|
| 1.0668 : 1939.1376 : 0.1524 | |
| index_units | m |
| code | None |
| description | Gamma-Ray |
| log_type | None |
| api | None |
| date | 10-Oct-2007 |
| null | -999.25 |
| run | None |
| service_company | Schlumberger |
| Stats | |
| samples (NaNs) | 12718 (0) |
| min mean max | 3.89 78.986 267.94 |
| Depth | Value |
| 1.0668 | 46.6987 |
| 1.2192 | 46.6987 |
| 1.3716 | 46.6987 |
| ⋮ | ⋮ |
| 1938.8328 | 92.2462 |
| 1938.9852 | 92.2462 |
| 1939.1376 | 92.2462 |
We can block this log based on some cutoffs:
gr_blocky = gr.block(cutoffs=[40, 100])
gr_blocky.plot()
<AxesSubplot:title={'center':'GR'}, xlabel='gAPI'>
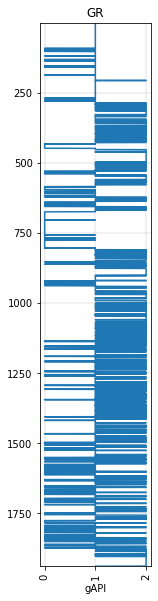
But now we’re not really dealing with regularly-sampled data anymore, we’re dealing with ‘intervals’. Striplog is sometimes a better option for representing this sort of data. So let’s try using that instead:
gr_blocky = gr.block(cutoffs=[40])
gr_blocky.plot()
<AxesSubplot:title={'center':'GR'}, xlabel='gAPI'>

We could find the thickest bed in this result using NumPy (i.e. the longest run of 1’s):
from itertools import groupby
samples = max(sum(run) for val, run in groupby(gr_blocky.values) if val)
samples * 0.1524
197.0532
But it’s much easier to use Striplog, which can compute the resut directly from the gr log itself. (You have to pass ‘components’ to the from_log() function, but it doesn’t matter what they are.
from striplog import Striplog
s = Striplog.from_log(gr, basis=gr.basis, cutoff=[40], components=[None, None])
s.thickest().thickness
197.05320000000017
Let’s look at how to do that with actual components…
Make a striplog instead of a blocked log#
Striplog objects are potentially a bit more versatile than trying to use a log to represent intervals.
We have to do a little extra work though; for example, we have to tell Striplog what the intervals represent… the contents (lithologies or whatever) of an interval are called ‘components’.
We also have to pass the depth separately, because Striplog doesn’t know anything about welly’s Curve objects.
from striplog import Striplog, Component
comps = [
Component(properties={'lithology': 'sandstone'}),
Component(properties={'lithology': 'siltstone'}),
Component(properties={'lithology': 'shale'}),
]
s = Striplog.from_log(gr, basis=gr.basis, cutoff=[40, 100], components=comps)
s
Striplog(853 Intervals, start=1.0668, stop=1939.1376)
You can plot this:
s.plot(aspect=3)

For a more natural visualization, it’s a good idea to make a legend to display things. For example:
from striplog import Legend
L = """comp lithology, colour, width, curve mnemonic
sandstone, #fdf43f, 3
siltstone, #cfbb8f, 2
shale, #c0d0c0, 1
"""
legend = Legend.from_csv(text=L)
legend
| width | component | hatch | colour | ||
|---|---|---|---|---|---|
| 3.0 |
| None | #fdf43f | ||
| 2.0 |
| None | #cfbb8f | ||
| 1.0 |
| None | #c0d0c0 |
Now the plot will look more geological:
s.plot(legend=legend)
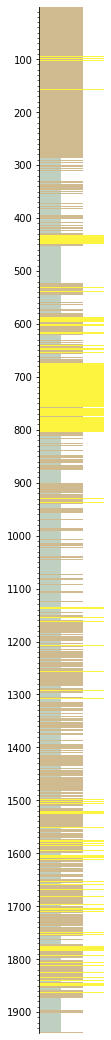
Let’s simplify it a bit by removing beds thinner than 3 m, then ‘annealing’ over the gaps, then merging like neighbours (otherwise you’ll likely have a lot of beds juxtaposed with similar beds):
s = s.prune(limit=3).anneal().merge_neighbours()
s.plot(legend=legend)
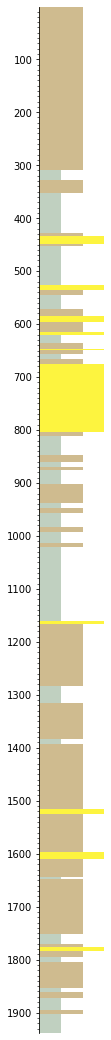
The easiest way to plot with a curve is like:
w.data['strip'] = s
tracks = ['MD', 'GR', 'strip']
C = """curve mnemonic, colour, width
GR, #ff0000, 1
GR-B, #ff8800, 1
"""
curve_legend = Legend.from_csv(text=C)
big_legend = legend + curve_legend
w.plot(tracks=tracks, legend=big_legend)
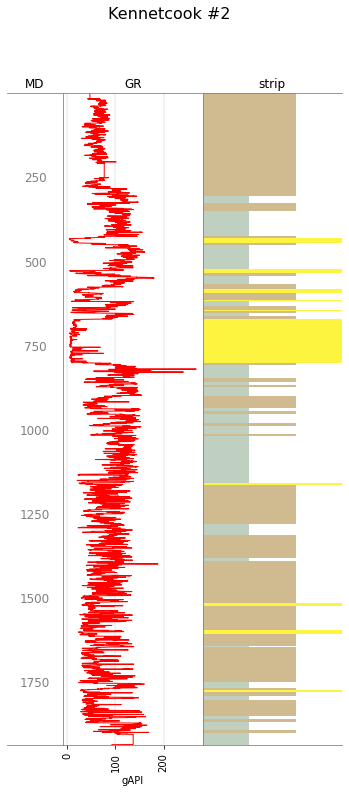
You can still make a blocky version from this Striplog:
gr_blocky, depth, comps = s.to_log(return_meta=True)
import matplotlib.pyplot as plt
plt.figure(figsize=(2, 10))
plt.plot(gr_blocky, depth)
plt.ylim(2000, 0)
(2000.0, 0.0)

Note that this is a NumPy array, because striplog doesn’t know about welly. But you could make a welly.Curve object:
from welly import Curve
gr_blocky_curve = Curve(gr_blocky, index=depth, mnemonic='GR-B')
gr_blocky_curve.plot()
<AxesSubplot:title={'center':'GR-B'}>

Let’s add this Curve to the well object w — then we can make a multi-track plot with welly:
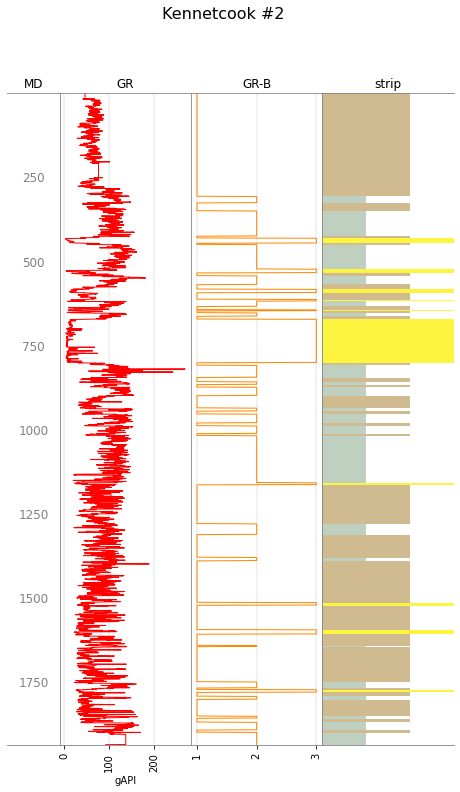
w.data['GR-B'] = gr_blocky_curve
tracks = ['MD', 'GR', 'GR-B', 'strip']
w.plot(tracks=tracks, legend=big_legend)
Blocking another log using these intervals#
Let’s use the intervals we just created to block a different log from the same well.
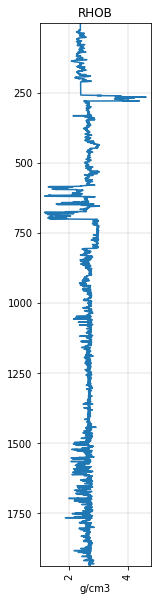
Here’s the RHOB log. We’ll block it using the intervals from the GR.
rhob = w.data['RHOB']
rhob.plot()
<AxesSubplot:title={'center':'RHOB'}, xlabel='g/cm3'>
Now we will ‘extract’ the RHOB data, using some reducing function — the default is to take the mean of the interval — into the intervals of the striplog s:
⚠️ Note that since v0.8.7 this returns a copy; before that, it worked in place.
import numpy as np
s = s.extract(rhob, basis=rhob.basis, name='RHOB', function=np.median)
Now each interval contains the mean RHOB value from that interval:
s[0]
| top | 1.0668 | ||
| primary |
| ||
| summary | 308.31 m of siltstone | ||
| description | |||
| data |
| ||
| base | 309.37199999999996 |
We can now turn that data ‘field’ into a log, as we did before. This time, however, we can keep the data as the value of the log — i.e. instead of having ‘bins’ from the cutoff (like 1, 2, 3, etc), we want the actual value from an interval (i.e. a density in units of g/cm3). To get this, we pass bins=False.
rhob_blocky, depth, _ = s.to_log(field='RHOB', bins=False, return_meta=True)
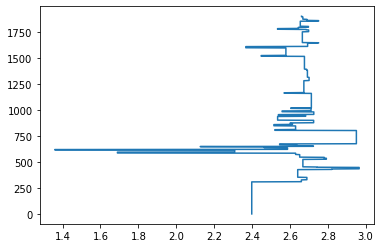
We can plot this…
plt.plot(rhob_blocky, depth)
[<matplotlib.lines.Line2D at 0x7fa9f4b1c730>]
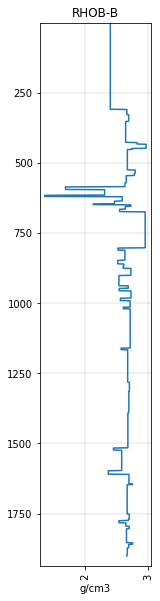
…but it’s more useful to make it into a Curve object and store it in the well object w:
rhob_blocky_curve = Curve(rhob_blocky, index=depth, mnemonic='RHOB-B', units='g/cm3')
w.data['RHOB-B'] = rhob_blocky_curve
rhob_blocky_curve.plot()
<AxesSubplot:title={'center':'RHOB-B'}, xlabel='g/cm3'>
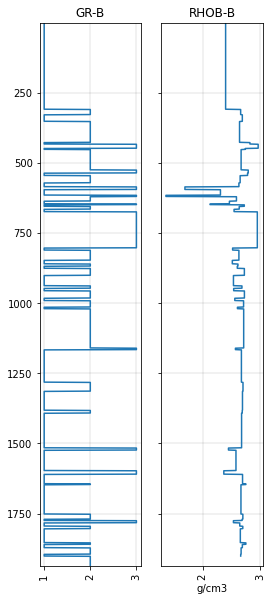
Now let’s plot it next to the blocky GR to verify that the blocks are the same:
fig, axs = plt.subplots(ncols=2, figsize=(4, 10), sharey=True)
gr_blocky_curve.plot(ax=axs[0])
rhob_blocky_curve.plot(ax=axs[1])
<AxesSubplot:title={'center':'RHOB-B'}, xlabel='g/cm3'>
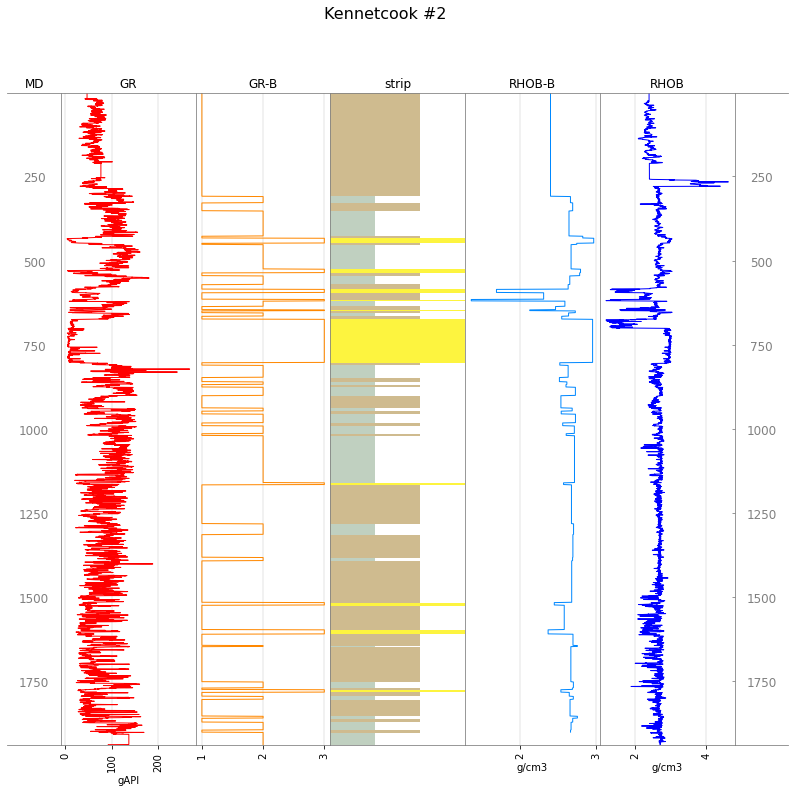
Or we can plot everything together using welly (if we first extend the legend again to accommodate the new logs):
C = """curve mnemonic, colour, width
GR, #ff0000, 1
GR-B, #ff8800, 1
RHOB, #0000ff, 1
RHOB-B, #0088ff, 1
"""
curve_legend = Legend.from_csv(text=C)
big_legend = legend + curve_legend
tracks = ['MD', 'GR', 'GR-B', 'strip', 'RHOB-B', 'RHOB', 'MD']
w.plot(tracks=tracks, legend=big_legend)
© 2020 Agile Scientific, licenced CC-BY.